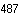Analysis of MeCP2 Mutation Frequency
Guide to the dynamic graphs
The graphs indicate the frequency of mutations at each amino acid. Moving the mouse over a column reveals information about mutations occurring at that amino acid. Click on a column to make the mutation information appear in a pop-up box. The information comprises the wild type amino acid, the amino acid residue number, a mutation description either indicating the new amino acid or describing the mutation in full, and the frequency of mutation. In cases of different mutations occurring at the same location, each mutation and their respective frequency is shown, separated by a slash ("/").
To rescale the graphs, type a number in the box labelled "scale".
The graphs are based on dynamic graphs seen at www.mecp2.org.uk, created by David Millband for The University of Edinburgh, Scotland.Mutation Classes
| Silent mutation: | A variation in exonic DNA that does not affect the amino acid sequence. |
| Missense mutation: | A point mutation that changes one amino acid into a different amino acid. |
| Nonsense mutation: | A point mutation changing an amino acid into a stop codon. |
| Frameshift mutation: | An insertion or deletion that changes all of the amino acids downstream of the mutation. |
| Miscellaneous mutation: | A mutation that does not fit into the above categories, or a compound mutation. |
| All mutations: | All mutations in the database. |
Mutation Frequency Charts
All mutationsSilent mutation
Missense mutation
Nonsense mutation
Frameshift mutation
Miscellaneous mutation
Chart of Mutation Frequency for All Mutations.
Move the mouse over the columns in the chart for further information.A guide to mutation graphs can be found here.
| |||||||
Chart of Mutation Frequency for Silent Mutations.
Move the mouse over the columns in the chart for further information.A guide to mutation graphs can be found here.
| |||||||
Chart of Mutation Frequency for Missense Mutations.
Move the mouse over the columns in the chart for further information.A guide to mutation graphs can be found here.
| |||||||
Chart of Mutation Frequency for Nonsense Mutations.
Move the mouse over the columns in the chart for further information.A guide to mutation graphs can be found here.
| |||||||
Chart of Mutation Frequency for Frameshift Mutations.
Move the mouse over the columns in the chart for further information.A guide to mutation graphs can be found here.
| |||||||
Chart of Mutation Frequency for Miscellaneous variations (Inframe variations, compound variations, etc.).
Move the mouse over the columns in the chart for further information.A guide to mutation graphs can be found here.
| |||||||